 The significance of mutations in the KIT and PDGFRA genes in the development and growth of GI stromal sarcomas (GISTs) is a topic that has been thoroughly discussed in the pages of this newsletter in an article by my colleague, Dr. Heinrich and in articles by other experts in the field. The Life Raft Group’s Science Coordinator, Jerry Call, has also made significant contributions on this topic. We know that mutations in KIT and PDGFRA occur early in GIST development, are key drivers of tumor growth and serve as the primary targets for kinase inhibitors like imatinib and sunitinib. In this article, I will concentrate on pragmatic issues related to testing GISTs for these mutations, with the goal of providing readers with an understanding of how testing is performed and what information it can (and cannot) provide.
The significance of mutations in the KIT and PDGFRA genes in the development and growth of GI stromal sarcomas (GISTs) is a topic that has been thoroughly discussed in the pages of this newsletter in an article by my colleague, Dr. Heinrich and in articles by other experts in the field. The Life Raft Group’s Science Coordinator, Jerry Call, has also made significant contributions on this topic. We know that mutations in KIT and PDGFRA occur early in GIST development, are key drivers of tumor growth and serve as the primary targets for kinase inhibitors like imatinib and sunitinib. In this article, I will concentrate on pragmatic issues related to testing GISTs for these mutations, with the goal of providing readers with an understanding of how testing is performed and what information it can (and cannot) provide.
As discussed in Dr. Heinrich’s article, genes are somewhat like books, each one divided into a series of exons (chapters) that are made up of codons (words), which in turn are made up of bases (letters). In GISTs, mutations can occur in any of 5 different exons of KIT or three different exons of PDGFRA. Additionally, the mutations vary from a single base change (substitution of one letter by another) to deletions of up to a dozen or more codons (words) in a row. Sometimes there is a combination of a deletion and a substitution, which is like deleting part of a sentence and altering the meaning of the remainder. The infamous KIT exon 9 mutation, for which there is evidence that a higher dose of imatinib may be appropriate, is caused by a duplication of two codons back-to-back, as though someone forgot to proofread the gene when it was copied during the routine division of a cell into two daughter cells.
Finding any of the above types of mutations within the 3.2 billion letters (bases) that comprise the library of DNA within a tumor cell is truly akin to finding a needle in a haystack. Fortunately, we have PCR (polymerase chain reaction) technology. This Nobel-prize winning approach to the analysis of DNA revolutionized molecular biology in the early 1980’s is now the mainstay in all laboratory testing for tumor mutations. The details of this approach are beyond the scope of this article, but one can think of PCR as a means of finding and Xeroxing selected chapters (exons) in any book (gene) in the DNA library. Considering that the DNA library is submicroscopic and quite fragile, this is quite a trick!
Before PCR can be employed, however, it is first necessary to extract and purify the DNA from a tumor. If fresh tumor tissue is available, this is a simple process that can be done in a couple of hours and yields high quality material for testing. Unfortunately, it is not customary to collect and store fresh tumor tissue outside of university hospitals. Instead tumor tissue is put into formaldehyde (to prevent it from rotting), then dehydrated and embedded into a block of wax (Figure 1). The purpose of creating such blocks is so that thin sections can be cut from them and mounted onto microscope slides (Figure 1). This 150-year old approach to the microscopic diagnosis of tumors may seem a bit dated, but it remains cost-effective and accurate, so pathology departments continue to use it worldwide. Given that nearly all GIST tissue available for mutation screening is embedded in paraffin blocks, it has become necessary for molecular testing laboratories to adopt methods that allow recovery of the tumor DNA from the wax.
This 150-year old approach to the microscopic diagnosis of tumors may seem a bit dated, but it remains cost-effective and accurate, so pathology departments continue to use it worldwide. Given that nearly all GIST tissue available for mutation screening is embedded in paraffin blocks, it has become necessary for molecular testing laboratories to adopt methods that allow recovery of the tumor DNA from the wax.
The standard protocol for extracting DNA from paraffin-embedded tumor tissue takes 24 to 72 hours, and involves the use of organic solvents. The resulting DNA is of poorer quality than that which can be recovered from fresh tumor tissue, but it still serves as a reasonable starting point for PCR-based testing of KIT and PDGFRA exons. Our laboratory has recently been experimenting with a new extraction protocol that may shorten the whole procedure to just 3 hours.
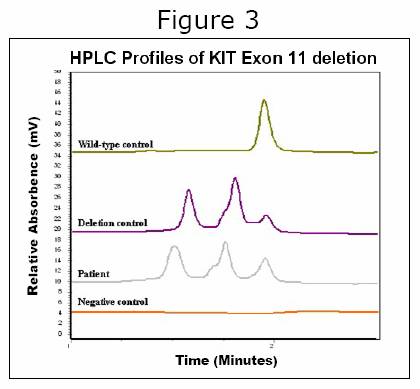
Once tumor DNA has been prepared and PCR reactions performed, the final step is to ‘read’ the amplified (Xeroxed) exons and look for a mutation. This can be done by several approaches. Many laboratories use standard DNA ‘sequencing’, which is essentially reading each exon line for line – a very laborious and time-consuming process. In the Heinrich & Corless laboratories, we adopted a different approach back in 2001 that has since been widely copied by other laboratories doing GIST analyses. Called ‘denaturing HPLC’, it is a method that allows us to ‘speed read’ each exon in 2.5 minutes, looking for any anomalies. This allows us to quickly dismiss the exons that are normal and focus on any that look anomalous (Figure 2). Such anomalies can then be pinned down by direct DNA sequencing to conclude the exact nature of any mutation that is present.
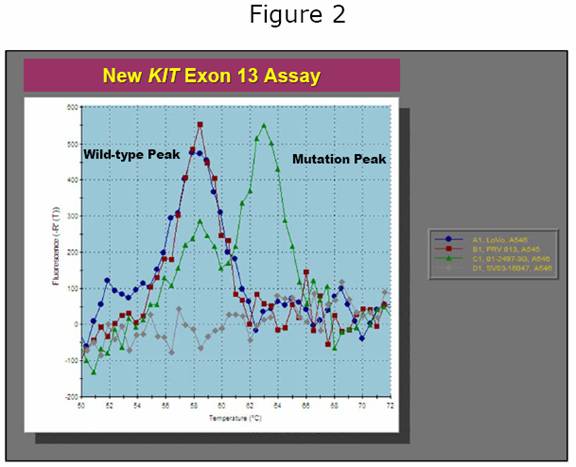 Denaturing HPLC is a powerful approach to mutation screening, but it is costly ($125,000 per instrument) and time-consuming to set up. For this reason, we have spent the past 18 months working on newer, faster approaches to mutation analysis. These approaches utilize a modified type of PCR in which special ‘probes’ that give off fluorescent light (like phosphorescence) are used to detect the presence of a mutation. An example of one such assay for the KIT exon 13 mutation is shown in Figure 3. The advantage of this assay is that it is very fast yet highly accurate. In combination with our new extraction protocol, we believe that this assay will permit exon 13 mutation screening to be done in a single day, whereas it currently takes 3 or 4 days. Similar assays will shortly be available for KIT exon 9 and PDGFRA exon 18 (including the imatinib-resistant D842V mutation).
Denaturing HPLC is a powerful approach to mutation screening, but it is costly ($125,000 per instrument) and time-consuming to set up. For this reason, we have spent the past 18 months working on newer, faster approaches to mutation analysis. These approaches utilize a modified type of PCR in which special ‘probes’ that give off fluorescent light (like phosphorescence) are used to detect the presence of a mutation. An example of one such assay for the KIT exon 13 mutation is shown in Figure 3. The advantage of this assay is that it is very fast yet highly accurate. In combination with our new extraction protocol, we believe that this assay will permit exon 13 mutation screening to be done in a single day, whereas it currently takes 3 or 4 days. Similar assays will shortly be available for KIT exon 9 and PDGFRA exon 18 (including the imatinib-resistant D842V mutation).
Along with our interest in finding primary KIT and PDGFRA mutations, our laboratories are examining secondary mutations that account for the onset of resistance to imatinib and more recently, sunitinib. These mutations are of keen interest to everyone concerned with imatinib resistance, which may develop after 12 to 36 months of treatment. One of the most common questions that I am asked is whether testing for resistance mutations is a good approach to deciding which drug should be used after imatinib. In theory, this is a good idea, because some mutations that cause resistance to imatinib are still sensitive to sunitinib, while others are resistant to both drugs but might respond to a newer agent.
 There are, however, two problems with testing for resistance mutations. First, it requires that a patient undergo a biopsy of one or more resistant GIST lesions in order to obtain the DNA needed for testing. Second, there is evidence emerging from a number of different laboratories that different tumor nodules may harbor different resistance mutations. More sobering still, work from our laboratories and that of Dr. Jonathan Fletcher using high-sensitivity assays for resistance indicates that even within a single tumor nodule there may exist up to three different resistance mutations, albeit at varying levels. What this implies is that once a GIST figures out a way to accumulate resistance mutations, it can do so quite prolifically. This undermines the significance of any particular resistance mutation that might be identified in a single biopsy. Indeed, basing treatment decisions on this approach might not be appropriate and could even prove harmful. For these reasons, we have decided not to offer routine testing for resistance mutations until more information from trials becomes available.
There are, however, two problems with testing for resistance mutations. First, it requires that a patient undergo a biopsy of one or more resistant GIST lesions in order to obtain the DNA needed for testing. Second, there is evidence emerging from a number of different laboratories that different tumor nodules may harbor different resistance mutations. More sobering still, work from our laboratories and that of Dr. Jonathan Fletcher using high-sensitivity assays for resistance indicates that even within a single tumor nodule there may exist up to three different resistance mutations, albeit at varying levels. What this implies is that once a GIST figures out a way to accumulate resistance mutations, it can do so quite prolifically. This undermines the significance of any particular resistance mutation that might be identified in a single biopsy. Indeed, basing treatment decisions on this approach might not be appropriate and could even prove harmful. For these reasons, we have decided not to offer routine testing for resistance mutations until more information from trials becomes available.
The concept that tumors should be subclassified by their mutation status is beginning to spread to other areas of oncology. Imatinib was first developed to treat chronic myelogenous leukemia (CML), a disease that is defined by a unique type of mutation. More recently, mutations in genes that are suitable targets for other imatinib-like drugs have been identified in lung cancer, breast cancer, thyroid cancer and endometrial cancer. Drug development for these targets is moving very quickly. So too, is the field of molecular testing, which may some day supplant the oldfashioned microscope and become the primary means for cancer diagnosis and drug assignment. In the meantime, selected use of molecular testing provides us with a valuable prism for understanding GIST biology and optimizing the treatment of these tumors.
Centers in the United States that offer KIT & PDGFRA gene mutation screening
•MD Anderson Cancer Center, Houston, TX
•ARUP Laboratories, Salt Lake City, UT
•Fox Chase Cancer Center, Philadelphia, PA
•Oregon Health & Science University, Portland, OR



