Introduction by Jerry Call
Today there are four major methods of determining risk of recurrence after initial surgery. These methods are pretty good at identifying low-risk and highrisk tumors. However, there is a fairly large group that fall between low-risk and high-risk. Those with so-called “moderate” or “intermediate” risk tumors are faced with the decision of whether or not to take Gleevec. New guidelines exist that recommend taking Gleevec for at least three years, with many experts advocating longer periods for highrisk patients.

Maria Debiec-Rychter, M.D.
Gleevec is an outstanding drug and a lifeline for the majority of GIST patients. However, it does have side-effects and these can be significant for some patients. In addition, like all cancer treatments, it is expensive. Thus it is extremely desirable to find better methods to determine which patients need to take Gleevec and which patients are essentially “cured” by surgery and don’t need to take Gleevec.
LRG Research Team member, Dr. Maria Debiec-Rychter describes three new genomic-based methods that appear to significantly outperform current methods of risk assessment:
- CINSARC
- AURKA expression
- Genomic Index
One of these methods, the CINSARC method, appears to have broader applicability to the wider sarcoma community as well. Importantly, the DNA CGHarray technique performed by Dr. Debiec-Rychter and colleagues can be performed from paraffin-embedded tumor tissue, the type of tissue that is already available/existing for most GIST patients.
Written by Maria Debiec-Rychter, M.D.
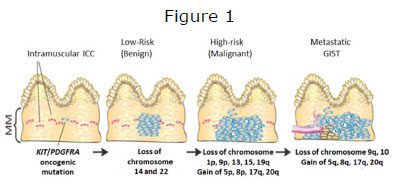
Figure 1. GIST progression is a multistep process that involves tumor initiation, tumor progression, and metastasis. A malignant GIST develops over time, as a result of an acquired mutation in KIT or PDGFRA and a series of sequential genomic abnormalities.
GISTs are heterogeneous tumors in terms of location, histology, molecular profile and prognosis. Although mutations of KIT or PDGFRA are early and most likely initiating tumorigenic events in GISTs, increasing evidence suggest that clinical behavior of GISTs are influenced by accumulation of other genomic/ chromosomal alterations (Fig. 1). Like in most tumors, a number of chromosomal changes occur during the progression of GIST from low risk to metastatic (Wozniak et al., 2007). By cytogenetic analysis approximately twothirds of GISTs demonstrate either total (monosomy) of partial loss of chromosome 14q. Loss of the long arm of chromosome 22 is observed in approximately 50 percent of GISTs. Loss of 1p and 15q are common and often coexisting, with higher frequency in high risk GISTs. Losses on chromosomes 1p, 9p (which harbours tumor suppressor genes CDKN2A and B), 9q, 10, 11p and 13q, and gains or amplifications on chromosomes 5p, 3q, 8q, and 17q are associated with malignant behavior (Fig. 2).
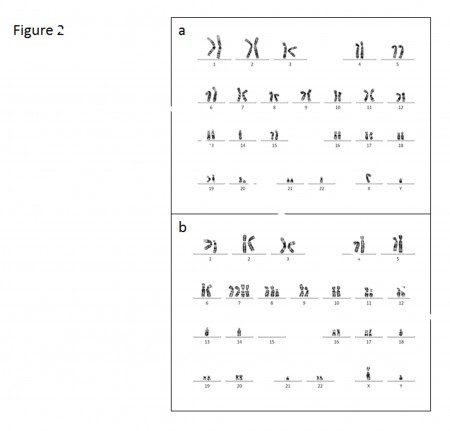
Figure 2. Chromosome abnormalities in low-risk GIST are relatively few compared with high-risk or overtly malignant tumors. Loss of chromosome 14 and 22 are two of the earliest cytogenetic changes that have been detected cytogenetically (A). The example of metastatic GISTs karyotype, characterized by complex chromosomal aberrations: losses of chromosome 1p, 13, 14, 15, 18, 21, gains of chromosomes 7 and 8q, and structural aberrations (arrows) (B).
Traditional methods of karyotyping have enabled rough analysis of chromosomal aberrations, but are imprecise and low-throughput. To map chromosomal changes with superior precision, researchers have turned to oligonucleotide-based comparative genomic hybridization (DNA array-CGH) (Fig. 3). By DNA array-CGH analysis, it was found that benign (more commonly referred to as very low- to low-risk) GISTs had a mean number of 2.6 aberrations per sample; malignant primary GISTs had a mean number of 7.5 aberrations per tumor, whereas the mean number of aberrations per metastatic GIST was 9 (El-Rifai et al., 2000; Wozniak et al., 2007). Genes targeted by these chromosomal changes and their contribution to clinical progression of GISTs are only partially known.
Alteration in DNA copy number is one of the many ways in which gene expression and function of cancer cells may be modified. In the past, several efforts have been made to establish biologic differences at the levels of gene expression profiles according to tumor’s KIT/PDGFRA genotypes, their anatomic site of origin and histopathologic grade of malignancy (lowrisk vs. intermediate vs. high-risk), based on the whole-genome expression profiling of heterogeneous cohorts of GISTs (Rubin et al., 2001; Antonescu et al., 2004; Ylipää et al., 2011). A variety of genes encoding oncogenes, tumor suppressors, proteins involved in cell and proteins involved in the control of cell motility have been found to be aberrantly expressed and/or amplified in GISTs.
In most expression profiling studies in sarcomas, the purpose has been to identify new diagnostic markers or to obtain better understanding of sarcoma pathogenesis. Only a few studies to date have tried to correlate expression profiles with the outcome of the disease. Recently, using a new methodological approach, Dr. Fredric Chibon and coworkers from the Bergonie Institute (France) have performed integrated genomic and expression profiling analysis of a large set of different sarcoma types, including 32 GISTs (Chibon et al., 2010).
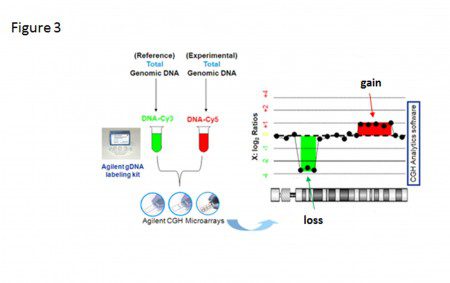
Figure 3.Schematic description of DNA array-CGH technology. In a typical array-CGH measurement, total genomic DNA is isolated from test (tumor) and reference (normal; usually blood white-cells) specimens, differentially labeled with fluorochromes and hybridized to DNA microarrays. The relative hybridization intensity of the test and reference signals at a given location is then proportional to the relative copy number of those sequences in the test and reference genomes. Data are typically normalized so that the modal ratio for the genome is set to some standard value, typically 1.0 on a linear scale or 0.0 on a logarithmic scale.
By analysing expression pro-files according to genome complexity (reflected by the number of chromosomal aberrations in a given tumour) and tumor’s histological grade (established by routine histopathological criteria), the researchers were able to identify a prognostic gene expression signature. This signature includes 67 genes involved with cell division (mitosis), chromosome management and, ultimately, genomic stability as a predictor of metastatic potential. The authors proposed the use of this system, which they termed CINSARC (Complexity INdex in SARcomas), as an adjunct to the current histologic grading system. Based on the system the tumors could be divided into two CINSARC classes of “good” or “poor” prognosis.
The results were compelling as the authors showed that CINSARC is more sensitive in assigning risk group than currently used procedures and can be applied to many tumor types including GISTs. Subsequently, through the collaborative efforts between French and Belgian sarcoma groups, these results were confirmed and extended by a more recent study, in which DNA array-CGH and expression profiling analysis was performed on 67 primary untreated GISTs (Lagarde et al., 2011). As a result not only the value of CINSARC signatures was validated (Fig. 4) but also several important correlates were found.
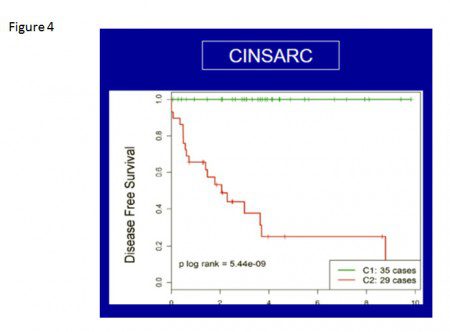
Figure 4. The correlation of disease progression of GIST and CINSARC signatures: the tumors were assigned according to CINSARC classes into two groups of “good” (C1, green) or “poor” (C2, red) prognosis.
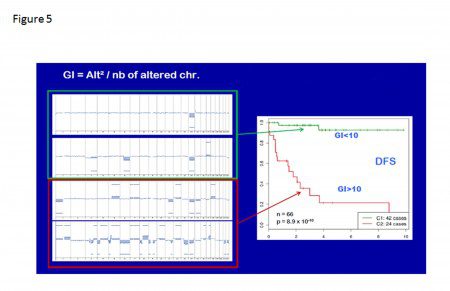
Firstly, the gene whose expression was most strongly associated with metastasis was AURKA (which maps to chromosome 20q), even though the AURKA locus was not amplified. Secondly, the deletion of the p16 (CDKN2A) and retinoblastoma (RB1) genes were identified, and these were mutually exclusive and likely causal events leading to increased AURKA and CINSARC gene expression, to chromosome rearrangement and ultimately to metastasis. In addition, the so-called Genomic Index that integrates the number and type of DNA copy number alterations was established. This index is a strong prognostic factor in GISTs (Fig, 5). The CINSARC class, level of AURKA expression and Genomic Index all outperform the currently used histopathologic grading system in determining the prognosis of patients with GISTs.

GISTs are the most frequent mesenchymal tumors of the gastrointestinal tract and are among the rare tumors to benefit from a targeted therapy. Thus, the development of a method for GIST prognostication has become essential for the proper clinical management of GIST patients, especially in the context of adjuvant treatment, where many patients are exposed to drug while only a small proportion will likely benefit from such treatment. In the past, many pathological criteria based on tumor site, tumor size, cell type, degree of necrosis and mitotic rate have been proposed for predicting the outcome of patients with GISTs. A consensus grading scheme based on tumor size and mitotic count was proposed by the US National Institutes of Health (NIH) in 2001 to estimate the prognosis of GIST patients.
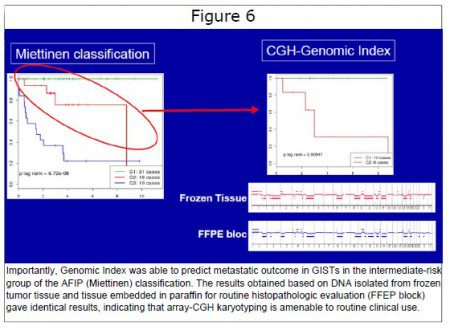
In 2006, the Armed Forces Institute of Pathology (AFIP) proposed an updated system taking into account also tumor location (Miettinen and Lasota, 2006). Both systems are based on histopathological assessment of tumor aggressiveness. Cut-off values defining risk groups have been determined empirically, but generate a large intermediate-risk group (approximately 25% of patients) for which adjuvant imatinib is controversial because the real metastatic risk is poorly defined. As mentioned above, high Genomic Index determined by array-CGH can identify poor prognosis patients in the group classified as intermediate-risk by the AFIP classification. (Figure 6) Array- CGH technique is already used in pathology laboratories using formalinfixed paraffin-embedded samples. Genomic array-CGH profiling could therefore be a powerful tool to manage imatinib therapy for intermediate-risk GIST patients.
Figure Notes Figure 1. GIST progression is a multistep process that involves tumor initiation, tumor progression, and metastasis. A malignant GIST develops over time, as a result of an acquired mutation in KIT or PDGFRA and a series of sequential genomic abnormalities.Figure 2. Chromosome abnormalities in low-risk GIST are relatively few compared with high-risk or overtly malignant tumors. Loss of chromosome 14 and 22 are two of the earliest cytogenetic changes that have been detected cytogenetically (A). The example of metastatic GISTs karyotype, characterized by complex chromosomal aberrations: losses of chromosome 1p, 13, 14, 15, 18, 21, gains of chromosomes 7 and 8q, and structural aberrations (arrows) (B).
Figure 3. Schematic description of DNA array-CGH technology. In a typical array-CGH measurement, total genomic DNA is isolated from test (tumor) and reference (normal; usually blood white-cells) specimens, differentially labeled with fluorochromes and hybridized to DNA microarrays. The relative hybridization intensity of the test and reference signals at a given location is then proportional to the relative copy number of those sequences in the test and reference genomes. Data are typically normalized so that the modal ratio for the genome is set to some standard value, typically 1.0 on a linear scale or 0.0 on a logarithmic scale.
Figure 4. The correlation of disease progression of GIST and CINSARC signatures: the tumors were assigned according to CINSARC classes into two groups of “good” (C1, green) or “poor” (C2, red) prognosis.
Figure 5. To take into account the number and the type of changes, a Genomic Index (GI) was calculated for each tumor profile as follows: GI= A² / C, where A is the total number of alterations (segmental gains and losses) and C is the number of involved chromosomes. The proportion of metastatic cases increased with GI. Stratification by GI at a cut-off of 10 split the tumors into two groups with very different outcomes.
Figure 6. Importantly, Genomic Index was able to predict metastatic outcome in GISTs in the intermediate-risk group of the AFIP (Miettinen) classification. The results obtained based on DNA isolated from frozen tumor tissue and tissue embedded in paraffin for routine histopathologic evaluation (FFEP block) gave identical results, indicating that array-CGH karyotyping is amenable to routine clinical use.
Glossary
Sarcoma: Malignant tumor originating from mesenchymal type of cells.
Karyotype: A picture of all the chromosomes from an individual’s cells. This is a test used to check for chromosome abnormalities. A picture of a person’s chromosomes is created by staining the chromosomes with a special dye, photographing them through a microscope and arranging them in pairs. It gives information about the number of chromosomes, the structure of their chromosomes and the sex of the individual. The normal human karyotypes contain 22 pairs of autosomal chromosomes and 1 pair of sex chromosomes. Short arm of chromosome is named p and long arm q.
Deletions: The loss of genetic material. The deletion can be heterozygous (copy number of 1) or homozygous (copy number of 0). Deletions in tumor cells may represent the inactivation of a tumor suppressor gene, and may have diagnostic, prognostic, or therapeutic implications.
Gains: A copy number gain represents the gain of genetic material. If the gain is of just one additional copy of a segment of DNA, it is called a duplication. If there is one extra copy of an entire chromosome, it may be called a trisomy. When seen in tumor cells, they may have diagnostic, prognostic, or therapeutic implications.
Amplifications: Technically, an amplification is a type of copy number gain in which there is a copy number 10. In the context of ca
ncer biology, amplifications are often seen in oncogenes. This could indicate a worse prognosis, help categorize the tumor, or indicate drug eligibility.
Array comparative genomic hybridization: DNA array comparative genomic hybridization (DNA array-CGH) is an efficient approach to scanning the entire genome for alterations in DNA copy number.
References
El-Rifai W, Sarlomo-Rikala M, Andersson LC, Knuutila S, Miettinen M. DNA sequencecopy number changes in gastrointestinal stromal tumors: tumor progression and
prognostic significance. Cancer Res. 2000;60:3899-903.
Wozniak A, Sciot R, Guillou L, Pauwels P, Wasag B, Stul M, Vermeesch JR, Vandenberghe P, Limon J, Debiec-Rychter M. Array CGH analysis in primary gastrointestinal stromal tumors: cytogenetic profile correlates with anatomic site and tumor aggressiveness, irrespective of mutational status. Genes Chromosomes Cancer 2007;46:261-76.
Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA, Demetri GD, Fletcher CD, Fletcher JA. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118-21.
Antonescu,CR, Viale A, Sarran L, Tschernyavsky SJ, Gonen M, Segal NH, Maki RG, Socci ND, DeMatteo RP, Besmer P. Gene expression in gastrointestinal stromal tumors is distinguished
by KIT genotype and anatomic site. Clin. Cancer Res. 2004; 10, 3282-3290.
Ylipää A, Hunt KK, Yang J, Lazar AJ, Torres KE, Lev DC, Nykter M, Pollock RE, Trent J, Zhang W. Integrative genomic characterization and a genomic staging system for gastrointestinal stromal tumors. Cancer 2011;117:380-389
Chibon F, Lagarde P, Salas S, Pérot G, Brouste V, Tirode F, Lucchesi C, Aurélien de Reynies, Audrey Kauffmann, Bui B, Collin F, Guillou L, Terrier P, Axel Le Cesne, Sylvie Bonvalot, Vince-Ranchère D, Jean-Yves Blay, Françoise Collin, Leroux A, Coindre JM and Aurias A. Prediction of clinical outcome in sarcomas and multiple types of cancer based on CINSARC, a gene-expression signature related to genome complexity. Nat. Med. 2010;16(7) :781-787.
Lagarde P, Perot G, Kauffmann A, Brulard C, Dapremont V, Hostein I, Neuville A, Wozniak A, Sciot R, Schoffski P, Aurias A, Coindre JM, Debiec-Rychter M, Chibon F. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Clin. Cancer Res. 2011 Dec 13. [Epub ahead of print] Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med 2006;130:1466-1478