Introduction to mutations in GIST
 The cells in the human body each contain instructions for making the building blocks that make up the body. These instructions are contained in the DNA and are organized into genes; one gene for each protein, about 30,000 genes in all.
The cells in the human body each contain instructions for making the building blocks that make up the body. These instructions are contained in the DNA and are organized into genes; one gene for each protein, about 30,000 genes in all.
New cells are created when existing cells divide. Sometimes errors in copying the DNA occur during the cell division process. Usually, these errors are corrected, or the cell is destroyed so that the errors are not passed on to new cells. Sometimes however, errors (mutations) can be passed on to the new cell and each new generation of cells. If enough critical errors occur, the cell may stop following the rules that normal cells live by and the beginning stages of cancer can occur.
GIST was one of the first and most powerful examples in the cancer world that the mutated gene(s) driving the cancer is more important than the location of the tumor. Most of today’s cancer therapies are designed to address the results of gene mutations.
In GIST, the most common genes with mutations known or believed to be the driver mutations in GIST are:
KIT – ~75-80%
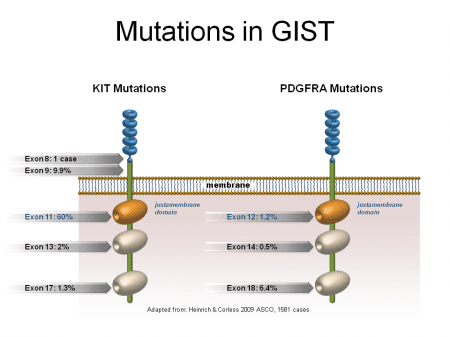
The information in genes is divided into different sections called exons and introns. Exons contain coding information and introns do not. Mutations in different exons in the gene cause changes in shape in different parts of the KIT receptor. Mutations in the following exons of the c-kit gene are known to occur in GIST.
Exon 11 – This is the most commonly mutated exon in GIST. Exon 11 mutations are found in about 60-65% of cases. Mutations in exon 11 generally respond to treatment with Gleevec better than mutations in other exons.
Exon 9 – Exon 9 mutations are the second most common mutation. Exon 9 mutations are found in about 10-12% of cases. In patients that have an exon 9 mutation, it occurs in the small bowel or colon about 98% of the time; however, they do NOT make up the majority of cases that occur in the small bowel or colon. In fact, about 2/3 of the cases that occur in the small bowel or colon are KIT exon 11 mutations and about 25% have exon 9 mutations. GISTs with exon 9 mutations have a lower response rate and a shorter response to standard dose Gleevec therapy when compared to exon 11 mutations. As a result a higher dose of Gleevec is generally recommended for patients with advanced/metastatic disease. They also seem to respond fairly well to Sutent.
Exon 13 (3%) and exon 17 (1%) mutations are rare in GIST. Primary exon 13 (typically K642E) mutations generally response well to imatinib, however primary exon 17 mutations are more often non-responsive to imatinib.
PDGFRA – ~8-10%
Some GIST tumor cells do not contain c-kit mutations. In about 5-8% of all GIST cases, a closely related gene, PDGFRA, is mutated. About 1/3 of the PDGFRA mutations may still respond to Gleevec and/or Sutent/Stivarga, but up to 2/3 of PDGFRA mutations do not respond to these drugs. These non-responsive mutations occur in one specific spot in exon 18 of the gene and are called a D842V mutation (PDGFRA mutation, exon 18, D842V). Several clinical trials for D842V inhibitors are in clinical trials for advanced/metastatic GIST with good to excellent early results.
KIT/PDGFRA WT1 – ~8-10%
Note: This GIST type has been commonly known as “wildtype GIST”, but is recently recognized as “No Other Specified” or NOS.
GIST tumors that do not have a mutation in KIT or PDGFRA have, in the past, been called “wildtype” GIST. Wildtype actually means that the gene in question is normal, so a “wildtype GIST” doesn’t really make sense. It was originally used to indicate that the KIT gene was normal and starting in 2003 (when PDGFRA mutations were discovered), the term wildtype GIST meant that a patient was tested for both KIT and PDGFRA mutations and none were found, thus, KIT and PDGFRA were “wildtype” or normal. This is often abbreviated as KIT/PDGFRA WT (wildtype).
Beginning in 2007, researchers have learned a lot more about this type of GIST. In fact, this group actually has a number of different driver mutations and it is no longer acceptable to stop testing after KIT/PDGFRA testing.
There are clinical implications for the 15% or so of GIST’s without KIT or PDGFRA mutations. These include drug selection, monitoring for additional types of tumors (SDH-deficient) and possible familial implications (SDH-deficient and NF1).
In most cases, if mutational testing is done, it stops after testing the KIT and PDGFRA genes, so if there is no mutation found in those two genes, the patient does not actually know their mutation. This is incomplete testing since as of 2018, we actually know quite a bit about the mutations these patients are likely to have. They include:
- SDH-deficient GIST – (SDHA, SDHB, SDHC, SDHD, SDHAF1, SDHAF2)
- NF1
- BRAF
- NRAS, KRAS or HRAS
- NTRK fusion mutations
- FGFR1
- And a number of other genes1.
Reference
- Nannini, M., Urbini, M., Astolfi, A., Biasco, G. & Pantaleo, M. A. The progressive fragmentation of the KIT/PDGFRA wildtype (WT) gastrointestinal stromal tumors (GIST). J. Transl. Med. 15, (2017). https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-017-1212-x
- Falchook, G. S. et al. BRAF Mutant Gastrointestinal Stromal Tumor: First report of regression with BRAF inhibitor dabrafenib (GSK2118436) and whole exomic sequencing for analysis of acquired resistance. Oncotarget 4, 310–315 (2013).
Next Steps
Article: Mutational Testing and Genetics
Knowing Your Mutation and Why It’s Important to Have Mutational Testing
Mutations and Mutational Testing FAQs
Treatments for GIST
References
- Demetri, G.D. et al. NCCN Task Force Report: Update on the Management of Patients with Gastrointestinal Stromal Tumors. Journal of the National Comprehensive Cancer Network 8, (2010).
- Casali, P.G., Blay, J.-Y. & On behalf of the ESMO/CONTICANET/EUROBONET Consensus Panel of Experts Gastrointestinal stromal tumours: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology 21, v98-v102 (2010).
- Pisters, P.W.T. et al. A USA registry of gastrointestinal stromal tumor patients: changes in practice over time and differences between community and academic practices. Ann Oncol (2011).doi:10.1093/annonc/mdq773.
